Transcriptomics: Lecture 2
Frontiers of Biotechnology: Bioinformatics and Systems Modelling
The University of Adelaide
stevie.pederson@thekids.org.au
The Kids Research Institute Australia
September 15, 2025
Helpful Links
I’d like to acknowledge the Kaurna people as the traditional owners and custodians of the land we know today as the Adelaide Plains, where I live & work.
I also acknowledge the deep feelings of attachment and relationship of the Kaurna people to their place.
I pay my respects to the cultural authority of Aboriginal and Torres Strait Islander peoples from other areas of Australia, and pay our respects to Elders past, present and emerging, and acknowledge any Aboriginal Australians who may be with us today
RNA-Seq
RNA Sequencing
According to Wang, Gerstein, and Snyder (2009)
RNA-Seq, also called RNA sequencing, is a particular technology-based sequencing technique which uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment, analyzing the continuously changing cellular transcriptome.
RNA Sequencing
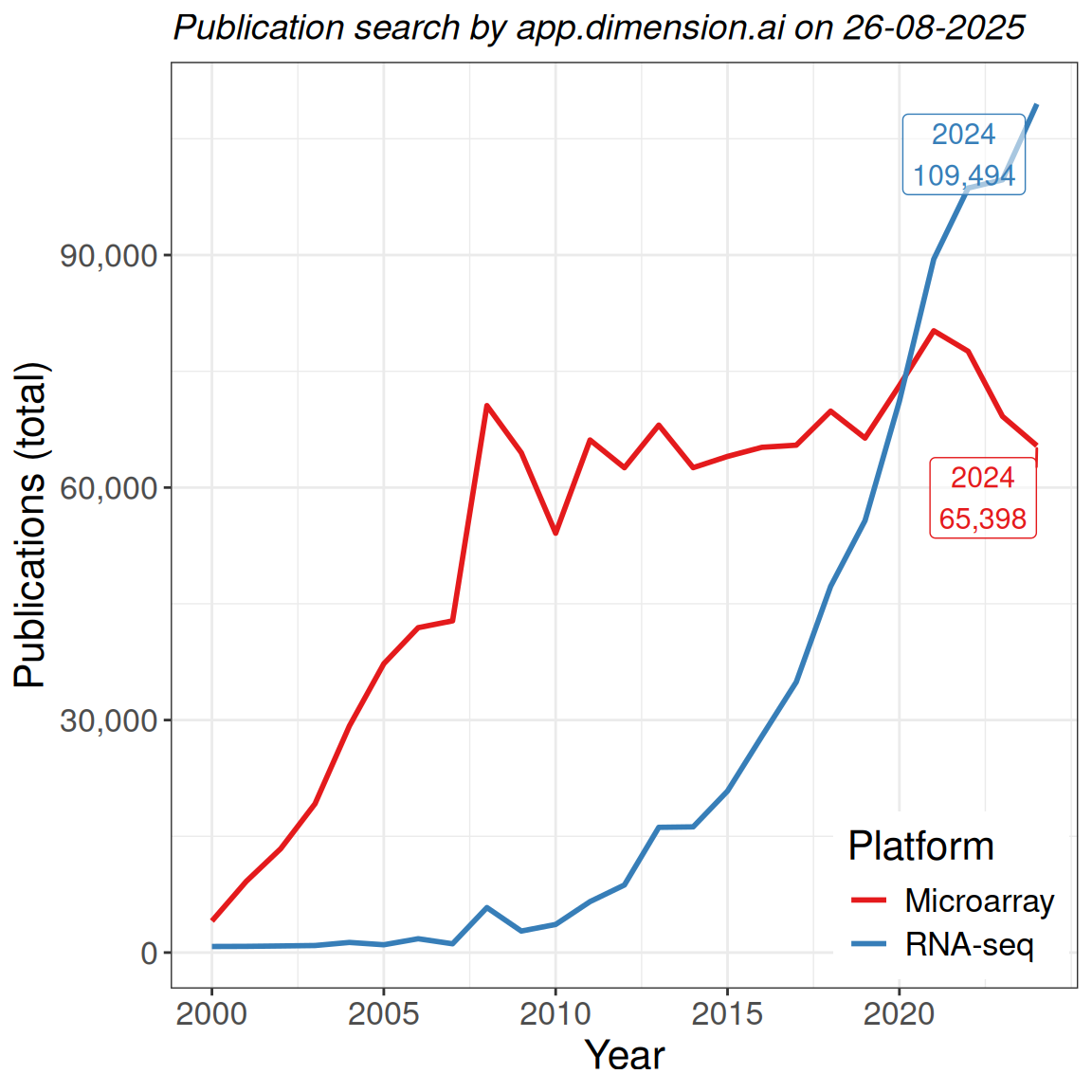
- Microarrays are still published regularly
- Also used extensively for methylation
- RNA sequencing is now the dominant technology
- Strong improvement for:
- transcript-level resolution
- un-annotated genes
- genomic variants
- allelic bias
RNA Sequencing
- Microarrays rely on probes for transcripts defined at design time
- Restricted in the number of transcripts/genes targeted
- Gencode 48 (GRCh38): 78,686 genes + 385,669 transcripts
- Probes capture non-specific binding
- Affymetrix use 25-mer probes
- Illumina arrays use 60-mer probes \(\implies\) sequences better targeted
These limitations do not exist for RNA-Seq
RNA Sequencing
- Directly sequence the biological material
- Map to most recent reference at any point in time
- Assemble a transcriptome (tissue specific)
- Detect InDels / SNPs in expressed sequences
- Multiple variations
- total-RNA or polyA transcripts \(\implies\) most similar to microarrays
- small-RNA libraries
- Long Reads (Oxford Nanopore, PacBio)
\(\implies\) originally isoform discovery, quantitative methods improving
The RNA Population Of a Eukaryotic Cell
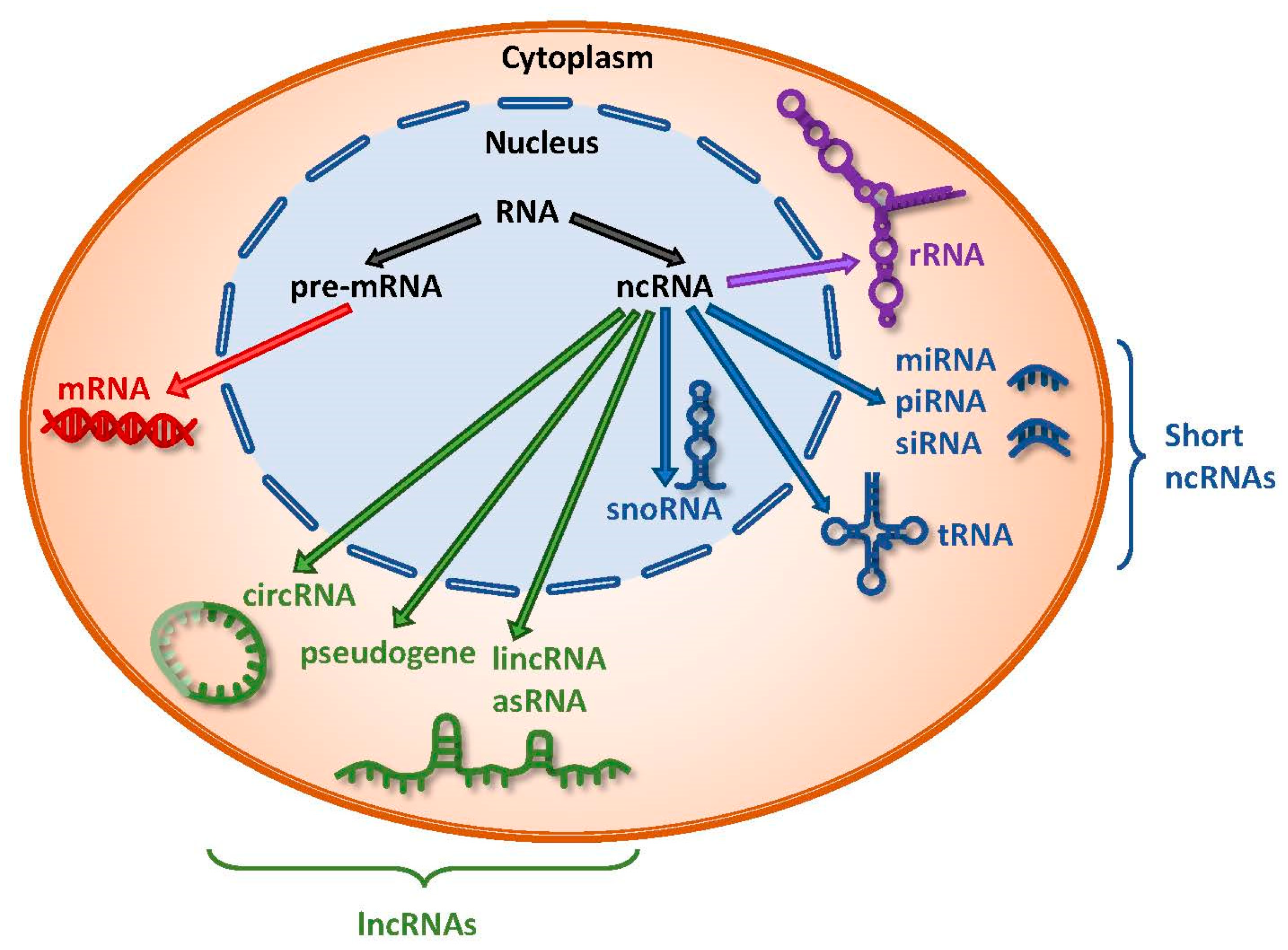
- rRNA \(\approx\) 80%1
- tRNA \(\approx\) 15%
- All other RNA \(\approx\) 5%
The Key Steps
- Focus from here on will be sequencing mRNA using short reads
- Library Preparation
- RNA Quality assessment
(i.e. RNA degradation) - Selecting target molecules
- Adding sequencing primers
- RNA Quality assessment
- Sequencing
- Alignment + Quantitation
- DE Gene Detection
- Downstream Analysis
- (Optional) Nobel Prize
RNA Selection
- Select for poly-adenylated RNA using oligo-dT-based methods
- Only extracts intact mRNA with a polyA tail (includes some ncRNA)
RNA Selection
- Enzymatically deplete rRNA sequences
- rRNA targeted using probes \(\implies\) dsRNA degraded
- Can additionally target hbRNA (whole blood)
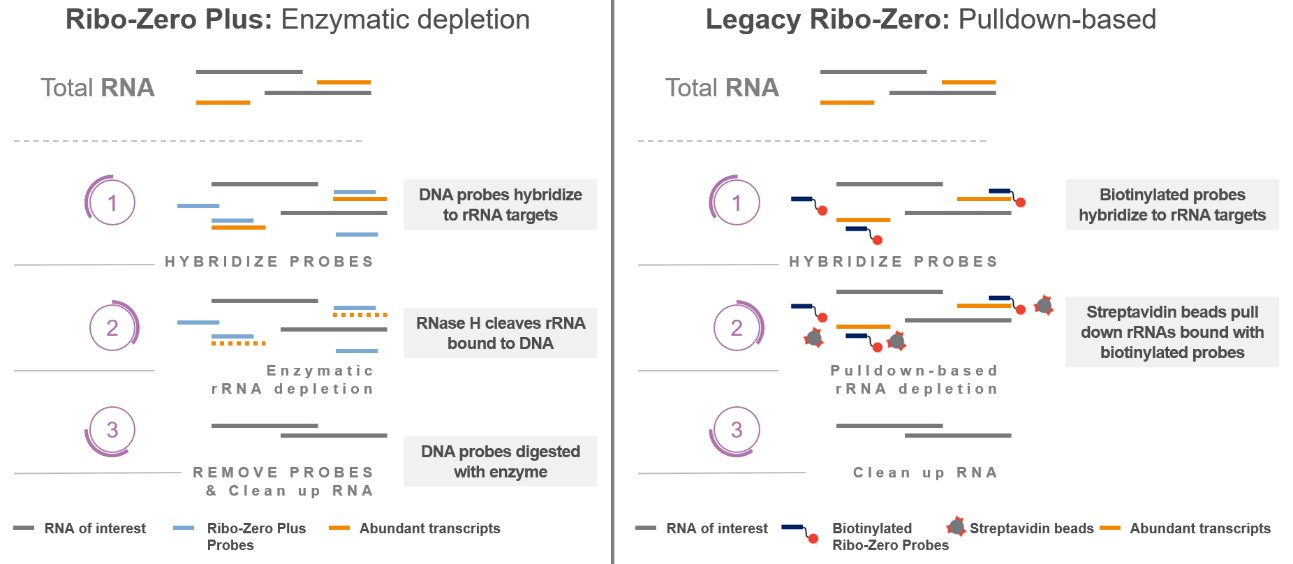
Image from https://support.illumina.com.cn
Library Preparation
- RNA is then fragmented and size selected (200-300nt)
- Very short transcripts always lost during this step
- cDNA produced
- Sequencing adapters added
- Indexes are unique to each individual library \(\implies\) always have replicates
- Optionally contain Unique Molecular Identifiers (UMI)
\(\implies\) Helps identify PCR duplicates
- Most RNA-Seq now retains strand-of-origin information (Stranded RNA-Seq)
- During PCR only the first cDNA template retained
Library Preparation
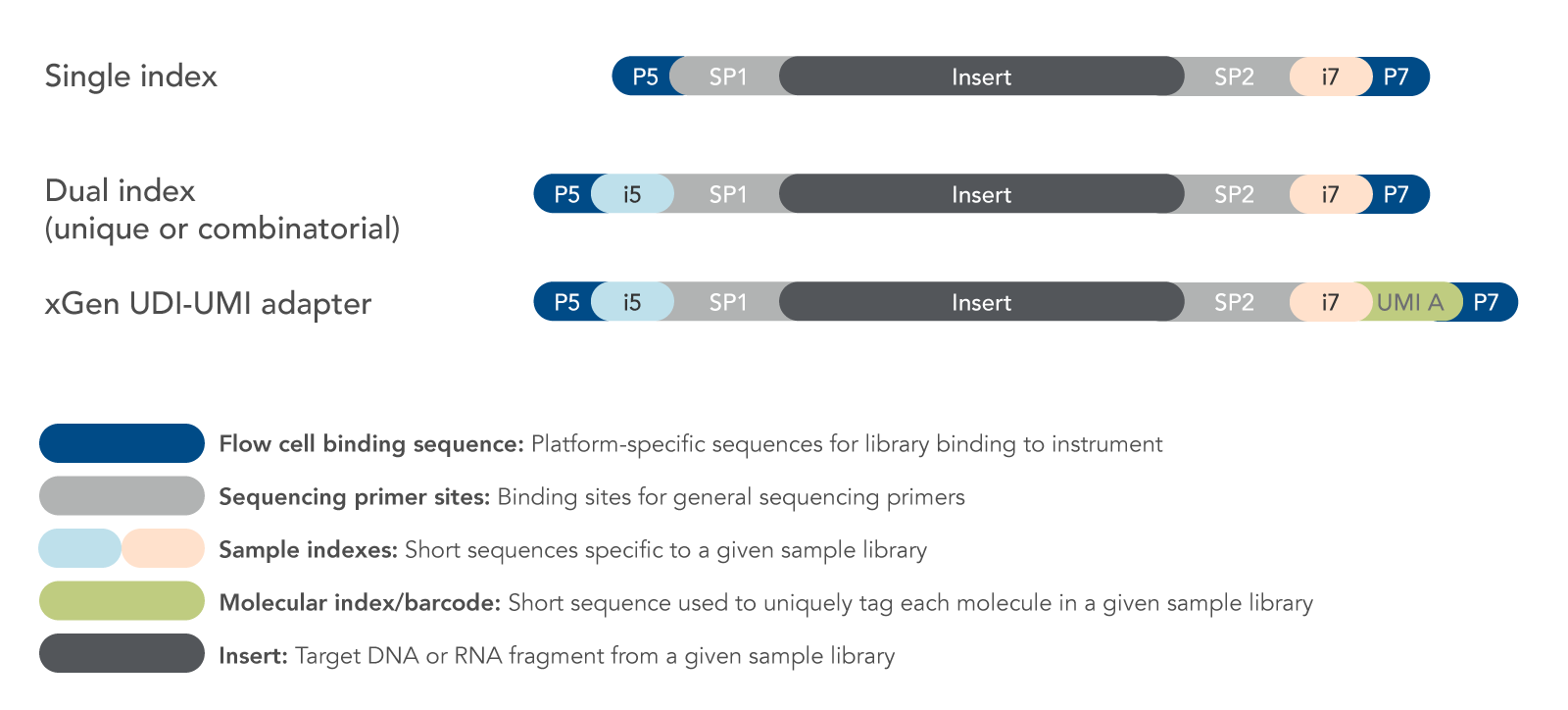
Image courtesy of https://www.lubio.ch/blog/ngs-adapters
Library Preparation
- Each double-stranded cDNA then has the final structure (+UMI)
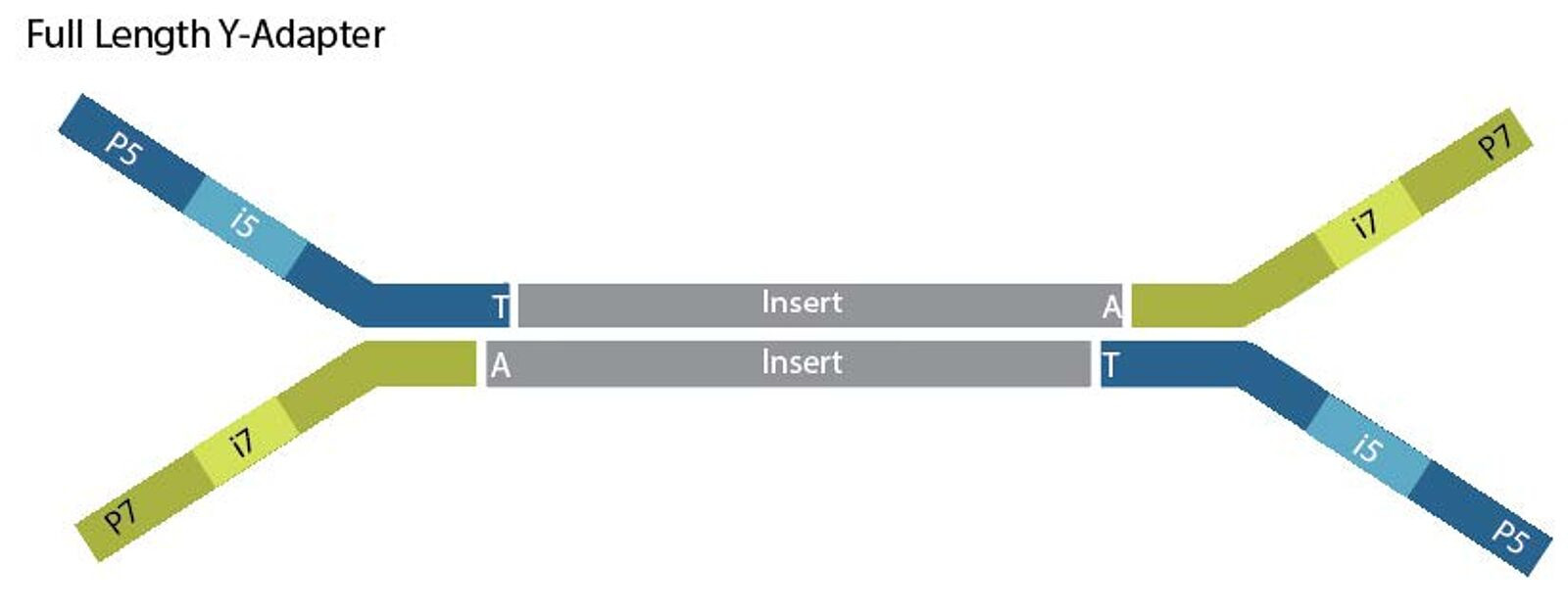
Image courtesy of https://www.lubio.ch/blog/ngs-adapters
Sequencing
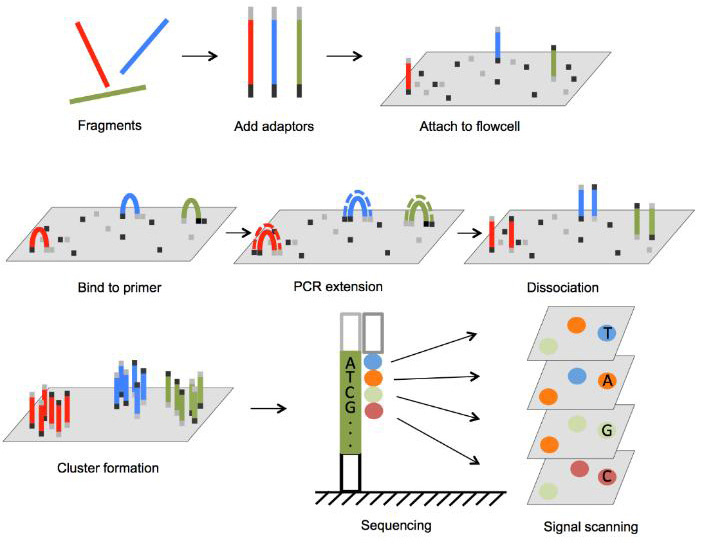
Alignment and Quantitation
Genomic Alignment
- Alignment to a reference genome requires a splice-aware aligner
- A GTF (Gene Transfer File) required when building the index
\(\implies\) Provides all exon-transcript-gene co-ordinates - New Gencode, Ensembl etc releases at regular intervals
- A GTF (Gene Transfer File) required when building the index
- Most common aligners are STAR (Dobin et al. 2013) & hisat2 (Kim et al. 2019)
- Return alignments as a
bamfile
- Return alignments as a
- Aligned reads are then counted to provide gene-level counts
- htseq (Anders, Pyl, and Huber 2014) and featureCounts (Liao, Smyth, and Shi 2014) are very common
- The same GTF should be used as during indexing
Genomic Alignment
Image from https://ucdavis-bioinformatics-training.github.io/2019_March_UCSF_mRNAseq_Workshop/data_reduction/alignment.html
Counting Alignments
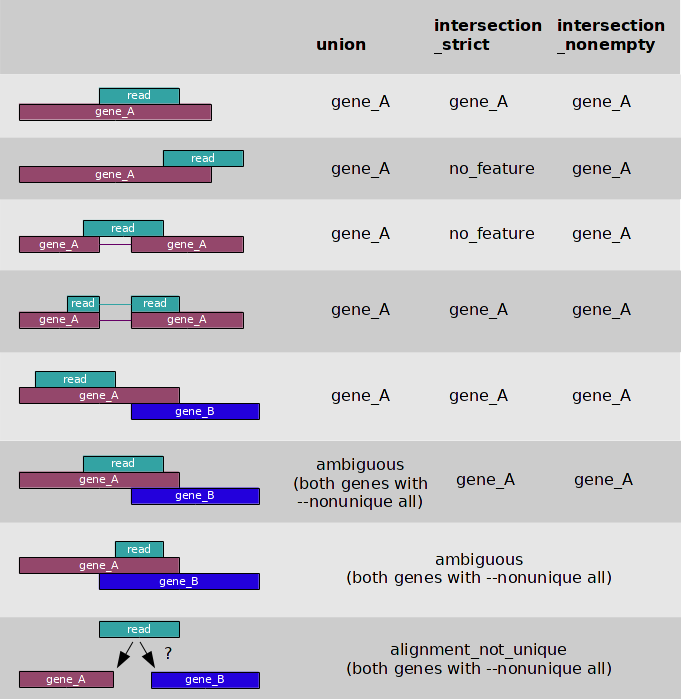
- Some alignments align beautifully within exon structures
- Some overhang a little
- Unspliced mRNA?
- Some genes are overlapping
- Stranded libraries can resolve
- Maybe bacterial reads span genes within an operon
Gene-Level Counts
- The region encoding a gene is (relatively) well defined
- An alignment within a gene is easy to assign to that gene
- Much more difficult to identify which transcript it came from
- Many transcripts share multiple exons
- Splice Junctions were the earliest approach
Gene-Level Counts
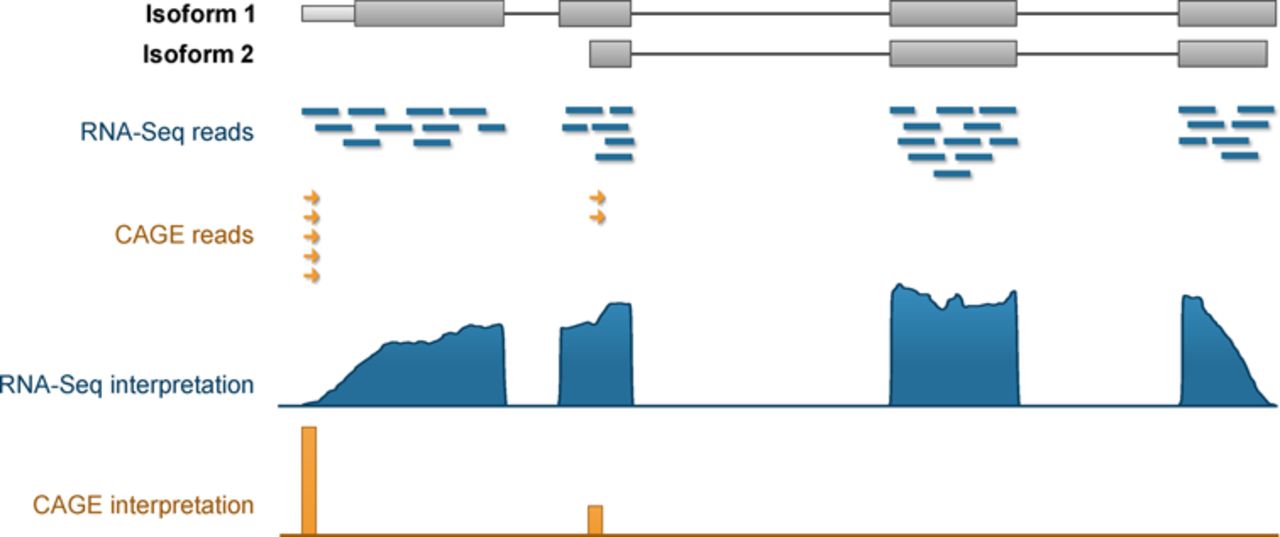
Image taken from Yu et al. (2015)
Transcriptome Alignment
- An alternative is to provide a reference transcriptome
- Alignments no longer need to be splice aware
- Reads can (& commonly do) align to multiple transcripts
- Much faster than traditional alignment
- Pseudo-alignment is used by kallisto (Bray et al. 2016)
- Statistically modelled expression estimates used by salmon (Patro et al. 2017)
- Return transcript-level counts without bam files
- Add transcript-level counts \(\implies\) gene-level counts
Pseudo-Counts
- Salmon counts are actually pseudo counts output by model fitting
- Predicts the proportion of library derived from transcript
- Fitted using EM-algorithm or Bayesian modelling
- Counts bootstrapped to provide uncertainty estimates of prediction
\(\implies\) Measures how confident we are in the transcript-level counts - Transcript-level counts can be scaled by uncertainty estimate (Baldoni et al. 2024) when performing DTE analysis
Differential Gene Expression Analysis
Differential Gene Expression Analysis
- A fundamental question:
Does the overall abundance of a gene differ between experimental conditions?
- Need a statistical approach to answer this question
Count-Based Data
- Under both reference-types \(\rightarrow\) counts represent expression
- These are discrete data (i.e. not continuous values)
- Microarrays were continuous values (fluorescence intensity)
- Modelled using log2-transformed values \(\implies \mathcal{N}(\mu, \sigma)\)
- Linear regression, \(t\)-tests etc
- Mean and variance are independent variables
- Count data is commonly modelled using a Poisson Distribution \(\implies \text{Poisson}(\lambda)\)
- Poisson variance is defined as being equal to the mean i.e. \(\sigma^2 = \mu\)
\(\implies\)Mean and variance are not independent variables
- Poisson variance is defined as being equal to the mean i.e. \(\sigma^2 = \mu\)
Count-Based Data
- In most cases the observed variance is greater than the mean\(\implies\) over-dispersion
- We use the Negative Binomial distribution to model counts for gene \(g\) in sample \(i\)
\[ y_{gi} \sim \text{NB}\left(\mu_{gi}, \sigma^2_g(\mu_{gi} +\phi\mu^2_{gi})\right), \text{where } \phi > 0 \]
- Can be thought of as like a Poisson with extra variation
- Extra variation is strictly defined in quadratic relationship to mean
- Sometimes described as a combination of technical & biological variation
Count-Based Data
- Fit generalised linear models (GLMs) to model counts and estimate logFC
- Implemented in edgeR (Chen, Lun, and Smyth 2016) and DESeq2 (Love, Huber, and Anders 2014)
- Slight differences in handling of over-dispersions
- Both default to FDR-adjusted \(p\)-values
- Same model for transcripts, but dividing counts by uncertainty estimate (Baldoni et al. 2024)
- Can also model transcripts as proportions of gene-level estimates
Count-Based Data
- Poisson/NB-GLMs fit the rate of an event, i.e. counts per fixed measurement window
- Sequencing data produces ‘libraries’ of counts \(\rightarrow\) total counts = library size
- In model fitting \(\rightarrow\) estimate rate as a function of library size
- TMM (edgeR) or RLE (DESeq2) approaches estimate scaling factors
- Moderates the effect of highly expressed genes which dominate library size
- In a perfect world \(\implies\) all scaling factors = 1
- Library size when modelling is multiplied by scaling factors
- The most effective normalisation methods for RNA-Seq
Count-Based Data
- A common alternative measure is counts per million (CPM) or logCPM
- CPM is simply counts divided by (library size / 1,000,000)
- No longer discrete \(\implies\) continuous data
- Relationship between mean and variance still retained for logCPM
- Can’t use naively in classic
limma-based linear models
- Can’t use naively in classic
- The limma-voom method (Law et al. 2014) uses weights to break the mean-variance relationship
- Can then assume normally-distributed data >\(\implies\) slightly more flexible models
- Alternatively, the
limma-trendworks effectively
Enrichment Testing
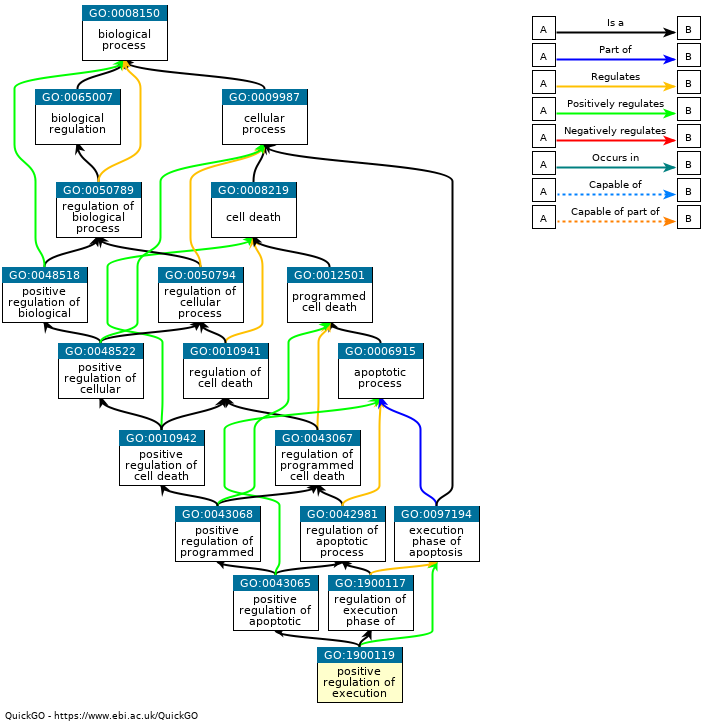
- Often want to identify key pathways associated with results
- Are genes from specific pathways or gene-sets most impacted
- Gene Ontology (GO) Terms:
- Carefully constructed, hierarchical database of terms
- Biological Process, Molecular Function and Cellular Component
Enrichment Testing
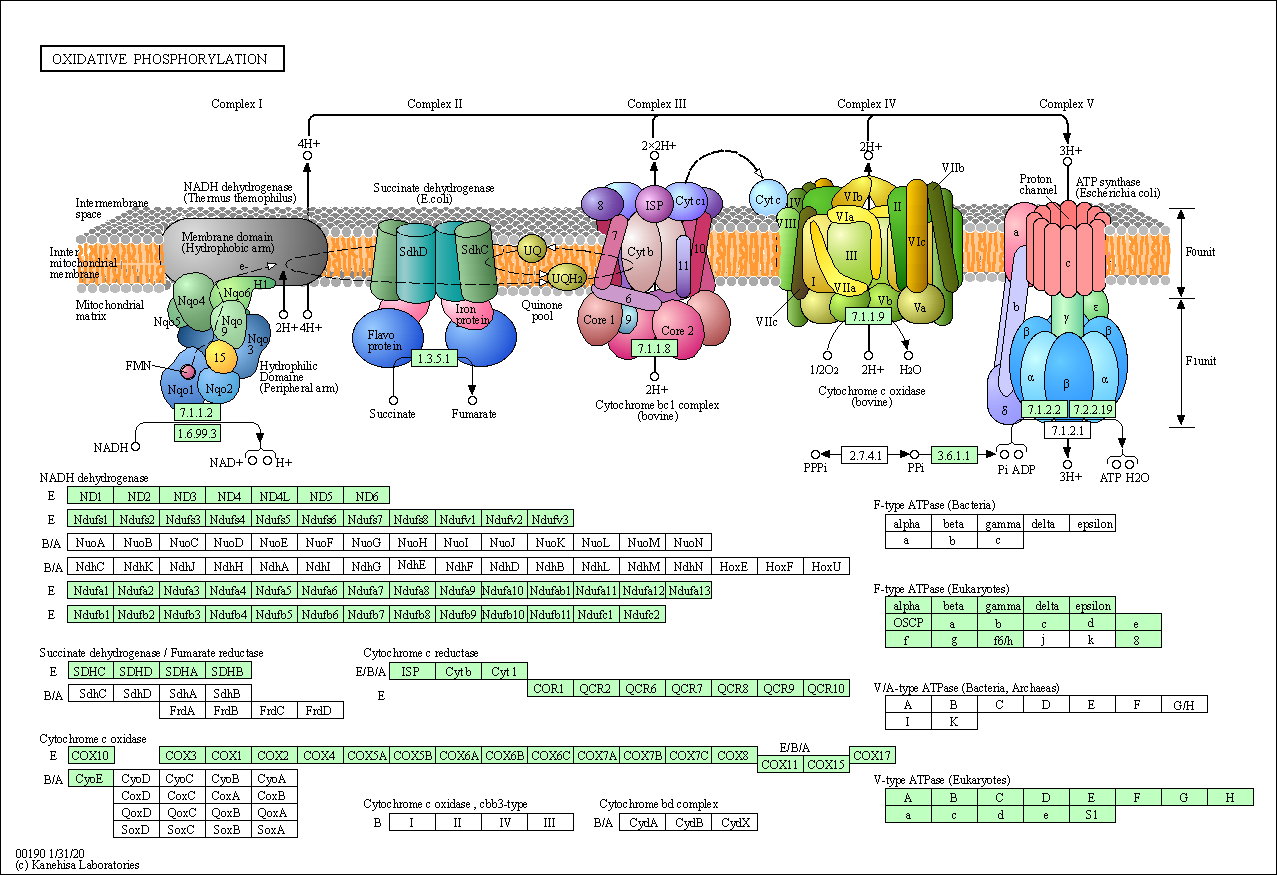
- Kyoto Encyclopedia of Genes and Genomes (KEGG):
- Molecular pathways with complete topology
Enrichment Testing
- Often use approaches based on Fisher’s Exact Test
- Compare enrichment of DE genes in a pathway vs not DE Genes in pathway
- Not-DE \(\equiv\) Background
- Pathways often share DE genes \(\implies\) biological expertise
- Multiple Testing across 1000s of pathways
| DE | Not DE | %DE | |
|---|---|---|---|
| In Pathway | 10 | 40 | 20% |
| Not In Pathway | 990 | 14000 | 6.7% |
- 0.28% not DE genes in pathway
- 1% of DE genes in pathway
Enrichment Testing
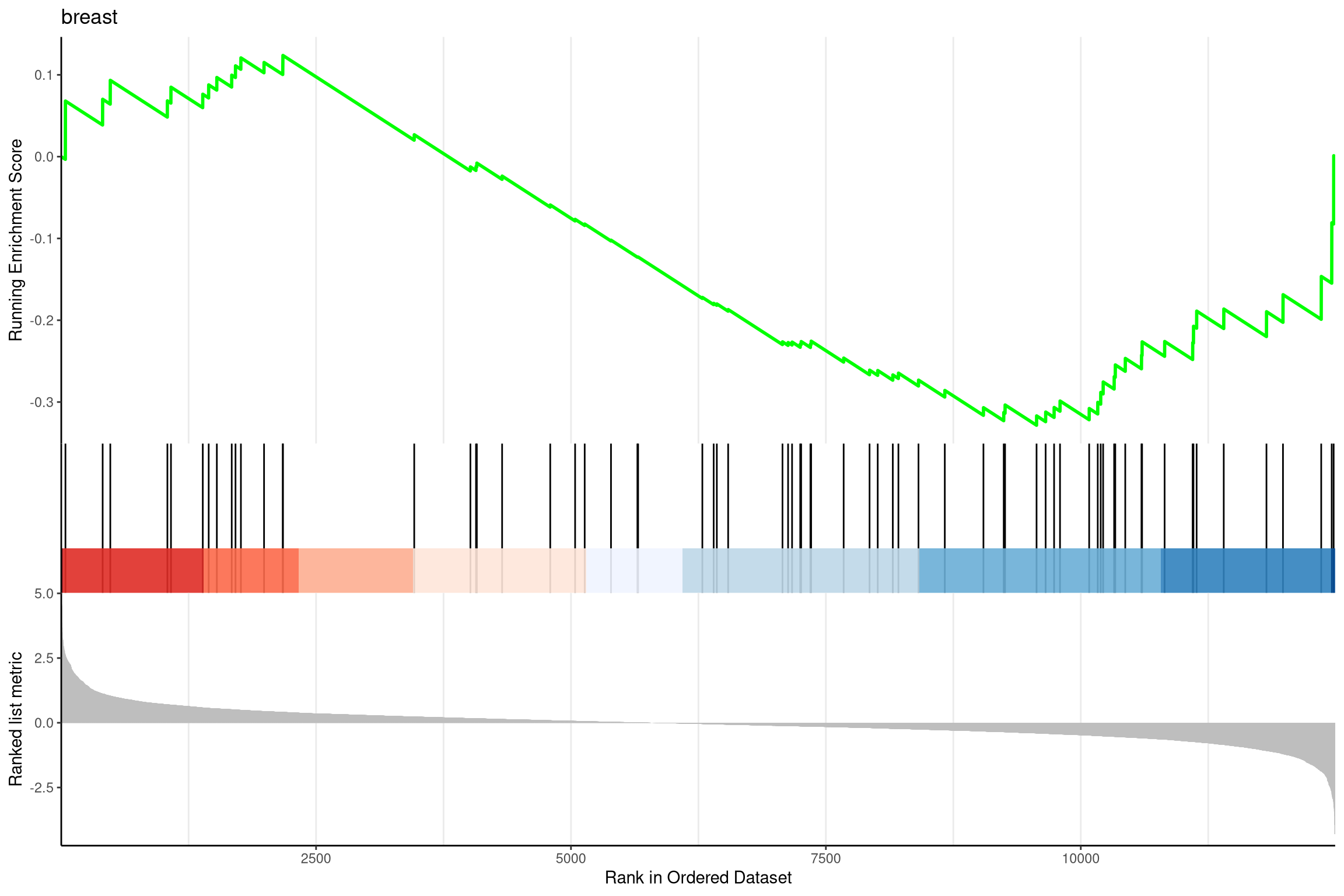
- An alternative approach is to use ranked lists
- Gene Set Enrichment Analysis (GSEA)
- No requirement to classify genes as DE or not DE
- Walk along the ranked list of genes increasing score if gene in gene-set
- In the example, begin at the right
- Largest deviation from zero is -ve
Beyond Differential Expression
Transcript Assembly
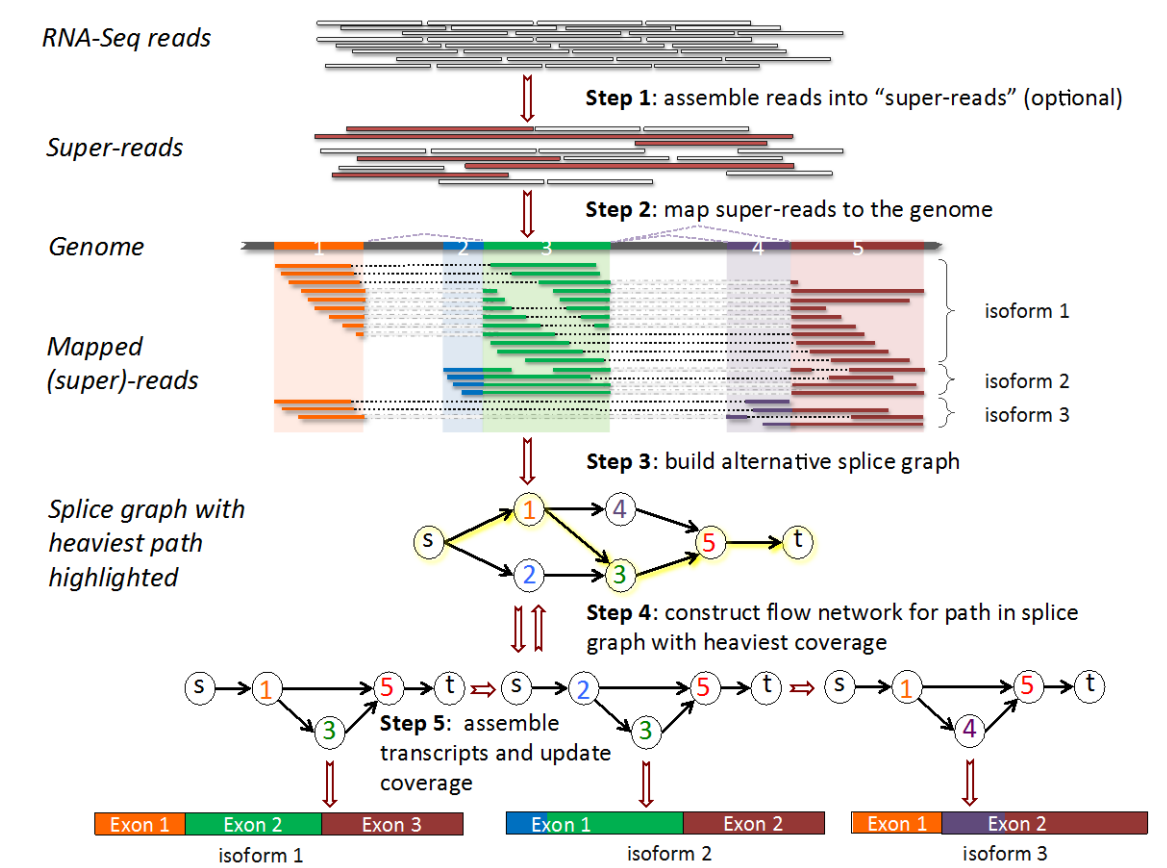
- With a good reference genome \(\implies\) StringTie can identify novel transcripts
- Un-annotated genes/lncRNA
Transcript Assembly
- StringTie returns a GTF with novel transcripts added to reference (See here)
- Gene & Transcript annotations capture the entire transcribed region
- Exons are annotated by transcript + gene
- Also returns counts for DE analysis
Transcript Assembly
- Complete de novo transcriptome assembly using Trinity (Grabherr et al. 2011)
- Can alternatively perform a full reference-guided assembly
- Also works with more distantly-related references
- Three basic modules
- Inchworm: Naively assembles reads into contigs
- Chrysalis: Pools contigs into de Bruijn graph
- Butterfly: Trims de Bruijn graph and compares against raw reads
- Check assembly against known conserved orthologs (BUSCO)
Transcript Assembly
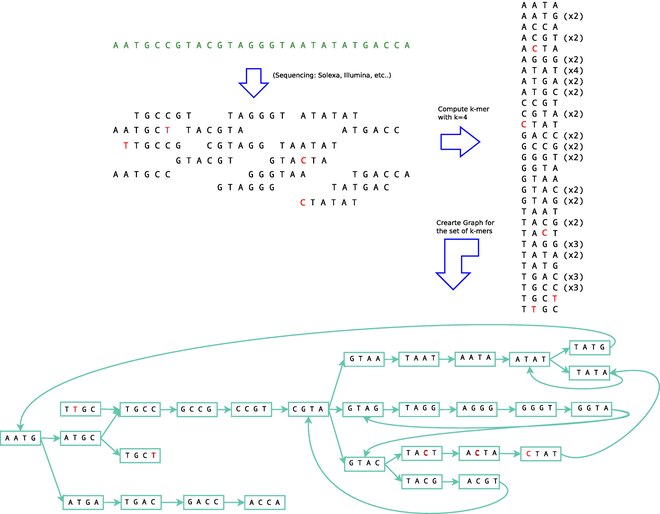
- Reads represent fragmented transcripts
- Reads broken into shorter k-mers
- Adjacent k-mers \(\rightarrow\) graph
- Wise choice of k helps extend graph well beyond read length
- Well-represented paths through graph \(\rightarrow\) transcripts
Transcript Assembly
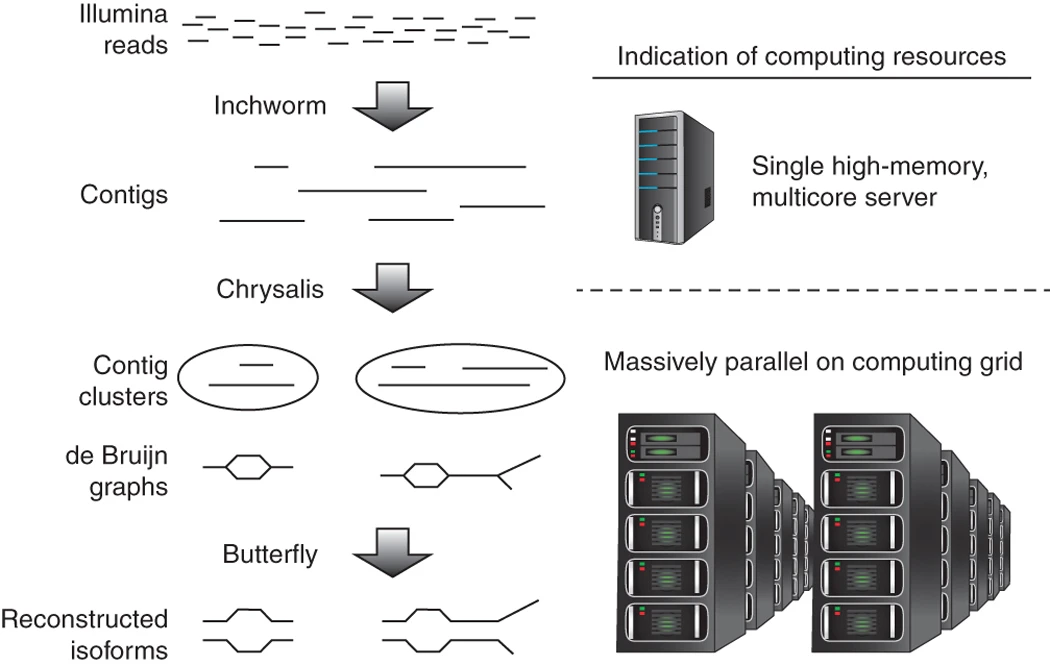
Figure from Haas et al. (2013)
Long Read Technology
- Most transcriptome assemblies performed using Trinity/StringTie
- Long Reads are now becoming the dominant platform
- Oxford Nanopore (ONT) reads from 50 bp to >4 Mb
- Pacific Biosciences (PacBio) up to 25kb
- Illumina maxes out around 2x150nt
- Both can sequence complete transcripts!
Long Read Technology
- Much more error prone
- Originally ~15% but now approaching Illumina error-rates
- Same molecule repeatedly sequenced
- Repeat sequencing makes quantitation more challenging
\(\implies\) Now becoming viable
High Resolution Technologies
Single-Cell Transcriptomics
- Single-Cell RNA-Seq is becoming a dominant transcriptomics platform
- Conventional RNA-Seq is now sometimes called bulk RNA-Seq
- Enables insights into highly heterogeneous samples (e.g. immune cells)
- Originally polyA-selected 3’ sequences \(\rightarrow\) full-length
- Incomplete transcript capture \(\implies\) many missing genes in each cell
Single-Cell Transcriptomics
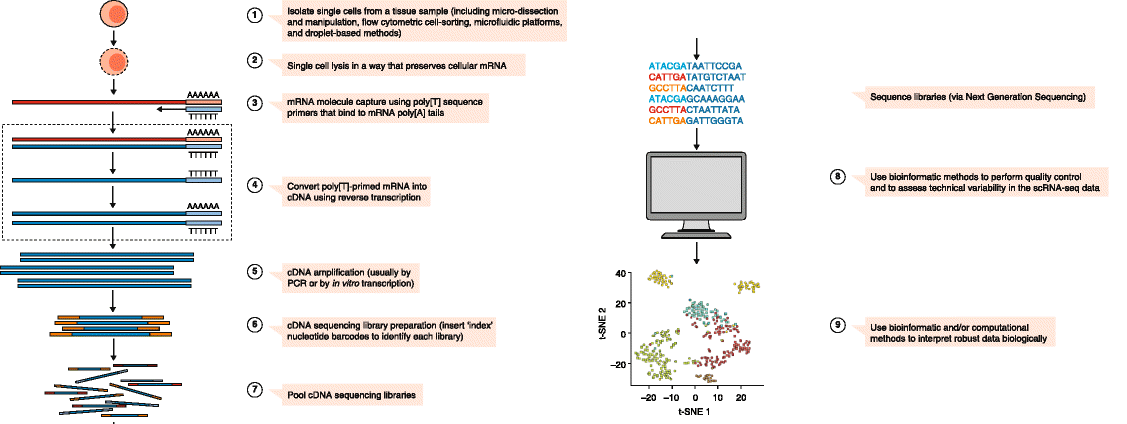
Figure taken from Haque et al. (2017)
Single-Cell Transcriptomics
- Single-Cell expression patterns used to cluster similar cells
- Clustering can be highly subjective
- Key marker genes identified to identify cell-types
- Often visualised using PCA, tSNE or UMAP to summarise expression values
- Clusters annotated as key cell-types
- Can be pseudo-bulked for DE comparison between cell-types within sample
- Methods such as CITE-Seq also label cells using tagged antibodies
- Cell types identified from cell-surface markers
Spatial Transcriptomics
- Spatial Transcriptomics uses tissue slices on a microscope slide
- Each spot on the slide contains oligo-dT probes with a unique spatial barcode
- Spatial barcode helps assign reads to a tissue region
- Interaction between key cell-types can be identified
- Some techniques use Fluorescence In Situ Hybridisation (FISH) to detect transcripts
- Cell boundaries detected in combination with classic stains
- Transcript location within the cell
- Can also cluster cell-types like scRNA then map back to tissue
- Multiple tissue layers possible \(\rightarrow\) $$$
Spatial Transcriptomics
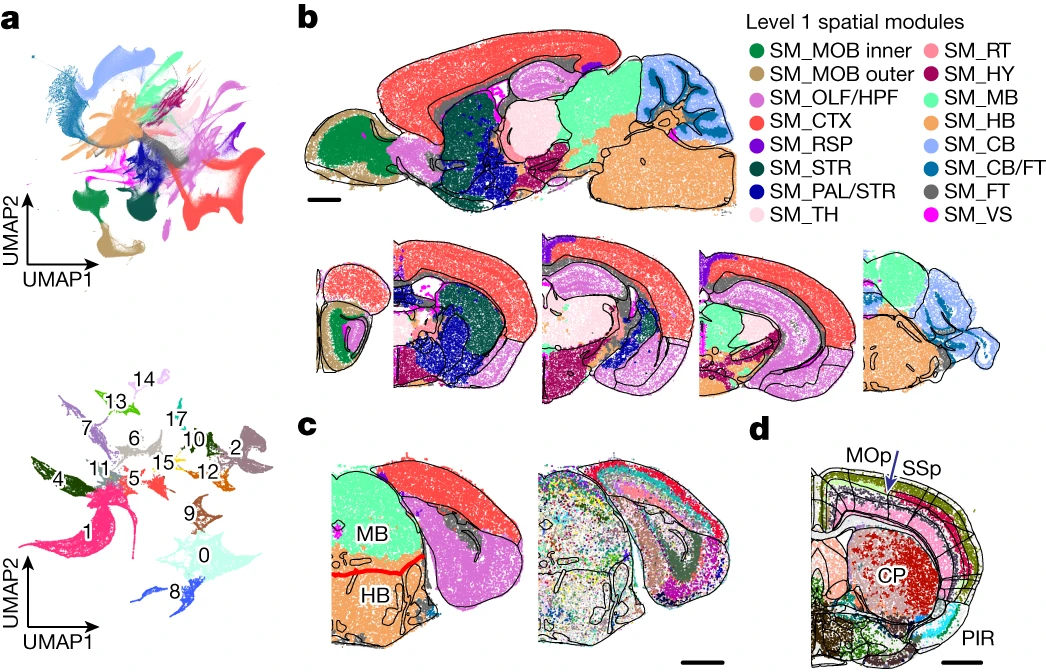
Figure from Zhang et al. (2023)


